NGS has not only revolutionized the field of genomics but also expanded its influence to various sectors including healthcare, agriculture, and environmental sciences, making it a cornerstone of modern biological research. Today, the landscape of genomic research is immensely diversified, with NGS technologies at the forefront driving innovations across multiple disciplines. In healthcare, for instance, NGS has enabled the detailed genetic profiling of tumors, facilitating the emergence of molecular tumor boards that tailor cancer treatments to individual genetic profiles, thereby enhancing the precision and effectiveness of oncology [2]. Similarly, in public health, NGS plays a crucial role in tracking the genetic diversity of pathogens like SARS-CoV-2, illustrating its critical function in managing health crises and shaping public health strategies [3].
The transformative impact of NGS extends its reach beyond medical applications into the broader realms of biological sciences. One significant area of impact is in the study of epigenetics, particularly how organisms respond to environmental stresses. For example, NGS has been instrumental in elucidating the epigenetic mechanisms that enable plants to adapt to adverse conditions, thereby informing strategies for crop improvement and sustainable agriculture [4]. Moreover, NGS has catalyzed emerging paradigms in genomic exploration, encouraging a holistic view of the life sciences. It integrates seamlessly with other 'omics' technologies, paving the way for systems biology approaches that promise to unravel complex biological networks and their interactions with the environment. This integrative approach is vital for the next generation of scientific breakthroughs, where complex biological questions require multifaceted strategies and deeper insights into genomic functions [2]. The ongoing development of NGS technologies continues to push the boundaries of what is scientifically possible, heralding a new era of genomic exploration that impacts not just scientific research but also practical applications in medicine, agriculture, and environmental management. These advancements have enabled genome-wide profiling of epigenetic modifications, which have significant implications for understanding stress responses in plants and improving crop sustainability [4].
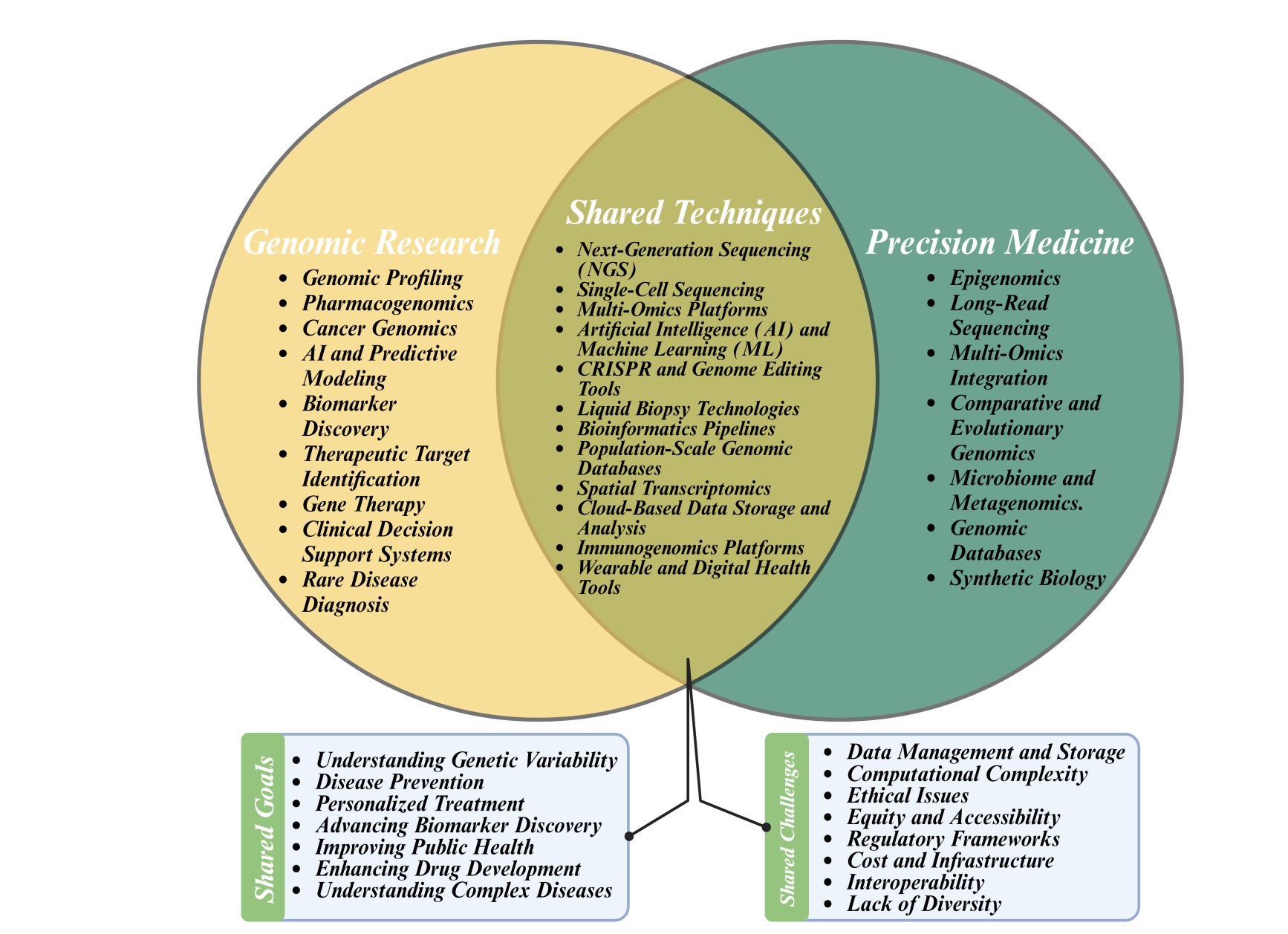
In this rapidly evolving field, the applications of NGS are vast and varied. From enhancing our ability to detect diseases at a molecular level to facilitating the exploration of genetic influences on human behaviour and physiology, NGS technologies are pivotal in driving forward the frontiers of genomic research [1]. They have revolutionized the detection of mutations and genomic variations in cancers, enabling precision medicine approaches tailored to individual genetic profiles [5]. The integration of NGS with emerging technologies such as artificial intelligence and machine learning is set to further enhance the speed and accuracy of genomic analysis. These integrations are already proving transformative in fields such as pharmacogenomics and evolutionary biology, providing a deeper understanding of genetic influences on health and disease [6]. Furthermore, the ability to sequence and analyse genomes at an unprecedented scale has opened new opportunities for studying microbial diversity, environmental sustainability, and ecological genomics [7]. The synthesis of key technological advancements, the current dynamic landscape, and the transformative impact of NGS form the bedrock of our manuscript, underscoring its foundational role in advancing genomic research while catalyzing broader societal and scientific developments. The potential for NGS to drive further innovations in understanding biological complexity and enhancing human life is immense. As these technologies evolve, they promise to deepen our understanding of life at a molecular level and bring forth new solutions to some of humanity's most pressing challenges [8].
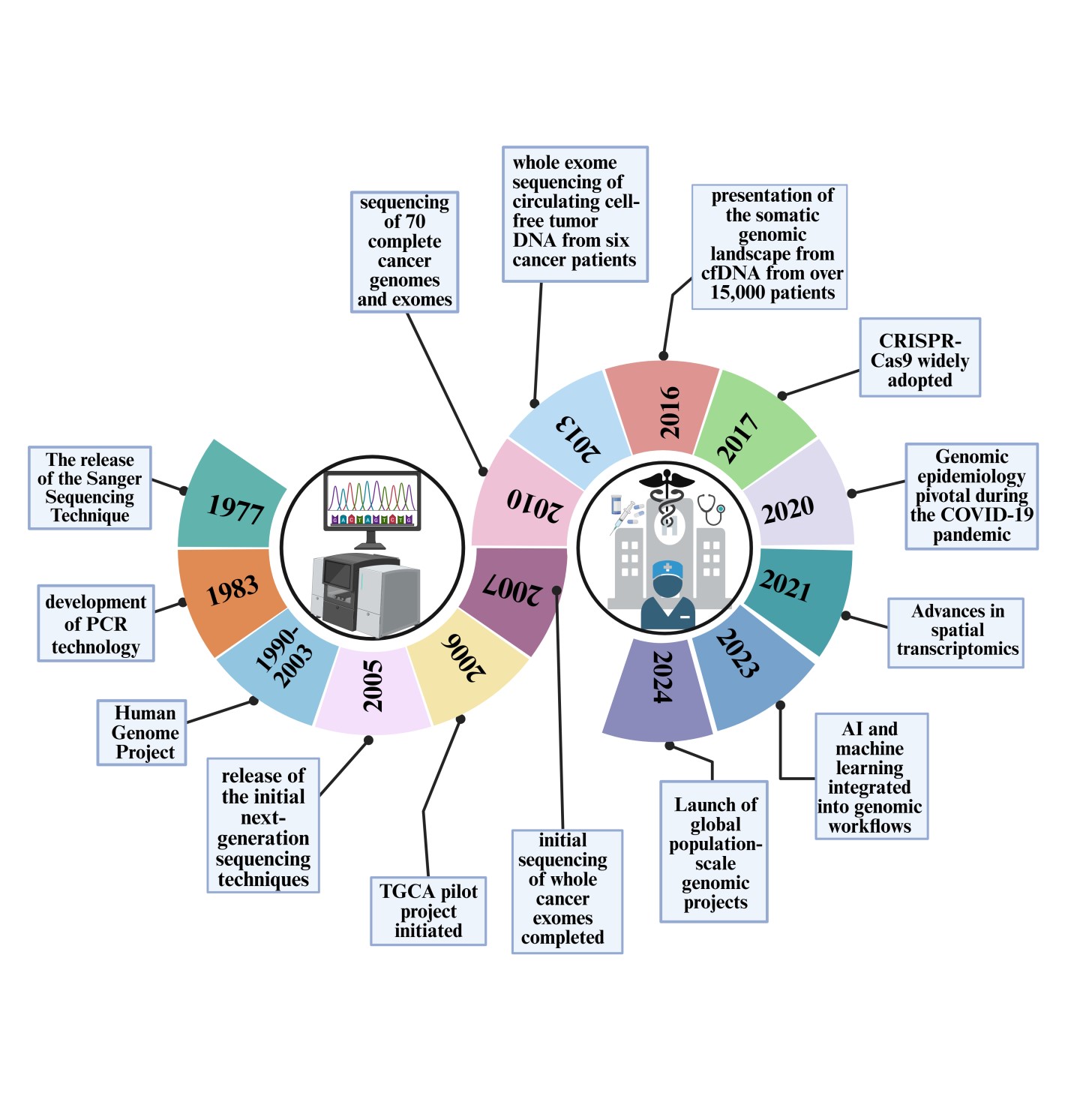
Technological innovations in sequencing platforms
The field of genomics has been revolutionized by technological innovations in sequencing platforms, particularly through the development of single-molecule sequencing technologies. These advancements allow for the real-time observation of DNA replication, providing unprecedented insights into the dynamics and regulatory mechanisms of genomic processes. For instance, Single-molecule Real-time (SMRT) sequencing has enhanced our understanding of epigenetic modifications and complex genomic structures that were previously challenging to resolve with older sequencing technologies [9]. Ultra-high-throughput sequencing approaches have further transformed genomic research by enabling the simultaneous processing of millions of DNA molecules. This capability has dramatically accelerated genomic data acquisition, reducing both the time and cost associated with genomic studies, thus broadening their application in medical research and personalized medicine [1]. The implications of these advancements are profound, impacting diverse areas from oncology to infectious disease research, where rapid genomic data generation is crucial for timely decision-making [2].